My Research Outline
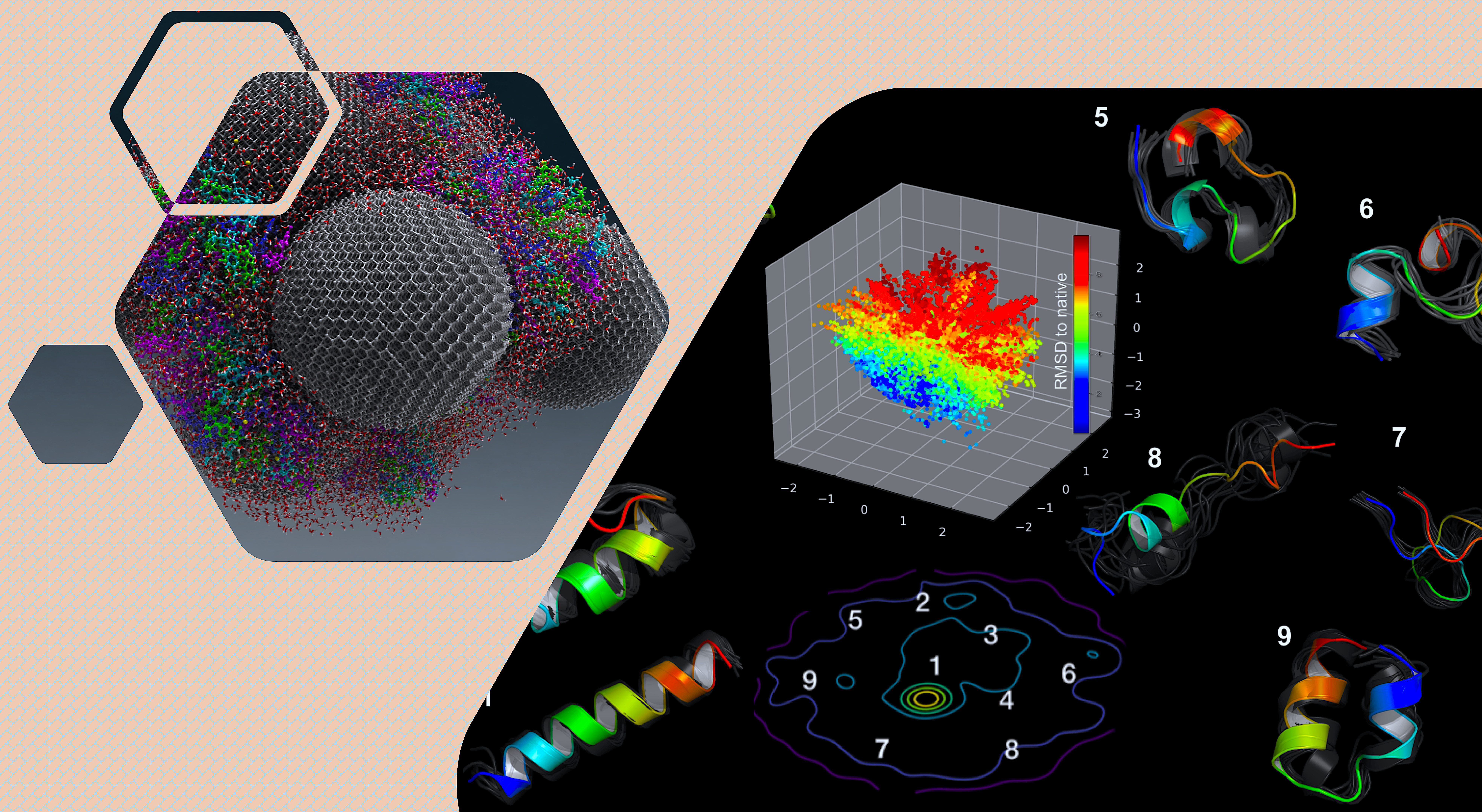
- General Overview: My current work lies on the interface of implementing Artificial Intelligence (AI) techniques, deploying multi-scale high performance accelerated simulations and performing scattering experiments (especially neutron and X-Ray) for problems related to
- soft matter systems especially Biomedical and biological sciences, and
- making new drug molecules with desired properties.
- Broader Goal:
- Optimal use of AI in integrative molecular approach: to find how the AI techniques can be optimally applied to the multi-scale modeling and simulation coupled with experiments
- Deriving fundamental Physics: to understand the underneath physics of bio(macro)molecular function, activity, folding, microscopic structure and dynamic behavior at different length and time scale and whether that knowledge could lead to designing new therapeutics.
- Reducing need for expensive computing and experiments by using AI: Additionally, developing AI tools to study large-scale datasets by learning the inherent hidden features of biomolecules in order to reduce the need for expensive computation or experiments.
- Sneak peek: Please take a sneak peek on my research at Projects section.
Interests
- Domain
- Biological/ Biomedical Science
- Complex/ Disordered system
- Polymer/ Polyelectrolytes
- Soft matter
- Proteomics
- Gene editing
- Techniques
- Artificial Intelligence, Deep Learning
- Simulation: specially Molecular Dynamics/ Atomistic/ Agent Based
- Modelling
- Data Analytics
- Scattering: specially Neutron/ X-ray/ Light